غير الغازية اختبار الوراثية قبل الولادة عن الشذوذ الجنين
[وهك] ممارسة نشرة وسريرية إدارة لمقدمي الرعاية الصحية.منح التعليمية التي تقدمها لصحة المرأة ومركز التعليم ([وهك]).
Most developed countries offer genetic screening and testing for aneuploidy as a standard part of prenatal care. Instead of invasive procedures such as chorionic villus sampling (CVS) or amniocentesis, definitive, noninvasive testing for fetal chromosomal abnormalities has long been the "holy grail" in obstetrics. If now appears practical to achieve prenatal genetic diagnosis using cell-free fetal DNA (cfDNA) in maternal blood. The ability to characterize cfDNA in maternal blood and the recognition of its potential was first appreciated 15 years ago (1). However, in June 2012 Kitzman and colleagues, in their report of non-invasive sequencing of the entire genome of an 18.5-week fetus using samples of maternal blood and paternal saliva, took the technology beyond detection of chromosomal or single-gene disorders to contemplation of genome-wide prenatal diagnosis of both recessive and dominant Mendelian disorders (2). The science of prenatal genetic diagnosis is moving at a pace that would have seemed unimaginable just a few years ago. Prenatal screening and diagnostic tests allow providers and parents to assess the health of a fetus, determine potential outcomes, plan for birth complications, decide whether to continue a pregnancy, and discover conditions that may affect future pregnancies. Science cannot yet fully obviate parental anxiety, but the increasing speed, specificity, reliability, predictive value, and integration of prenatal testing techniques can serve as effective anti-anxiety and pregnancy management tools. In this context, rapid, less invasive, broad-spectrum testing methods are increasingly recognized as particularly desirable. Prenatal DNA testing of a small sample of blood reduces the need for invasive procedures to detect Down syndrome and other genetic disorders. But this is only the beginning of what these tests will be able to do.
The purpose of this document is to discuss emerging clinical and scientific advances in the development of non-invasive prenatal testing for fetal genetic abnormalities. Known as fetal cell-free DNA testing (cfDNA), the test can detect Down syndrome and other chromosomal abnormalities through a sample of blood drawn from the mother. Recent review of the literature reveals non-invasive prenatal testing that uses cell-free fetal DNA (cfDNA) from the plasma of pregnant women offers tremendous potential as a screening tool for fetal aneuploidy. cfDNA testing should be an informed patient choice after pretest counseling and should not be part of routine prenatal laboratory assessment. cfDNA testing may not be offered to low-risk women or women with multiple gestations because it has not been sufficiently evaluated in these groups. A negative cfDNA test result does not ensure an unaffected pregnancy. A patient with a positive test result should be referred for genetic counseling and should be offered invasive prenatal diagnosis for confirmation of test results. Women's Health and Education Center (WHEC) recommends that women, regardless of maternal age, be offered prenatal assessment for aneuploidy either by screening or invasive prenatal diagnosis regardless of maternal age; cfDNA is one option that can be used as a primary screening test in women at increased risk of aneuploidy.
Introduction
The history of genetic diagnostic testing began in the mid-20th century. John Langdon Down first reported on the condition that came to bear his name in 1862. There were many theories as to the etiology of Down syndrome, but its true cause remained unclear for nearly a century until, in 1958, Dr. Jerome Lejeune employed recently developed chromosome banding analysis techniques (karyotyping) and observed and extra copy of chromosome 21 among individuals with Down syndrome. The condition was thereafter indentified as trisomy 21. The first clinical applications of karyotyping to prenatal testing specific to Down syndrome followed in 1966 (3). The 1980s brought widespread adoption of maternal serum analysis screening and individual risk estimation for Down syndrome. This decade also ushered in the detection of fetal structural anomalies via ultrasonographic scans that would become increasingly routine provide higher definition, and be performed into the third trimester. Karyotyping, or the microscopic examination of an individual's cultured-cell complete chromosome, achieved a reliably high degree of diagnostic accuracy. This became the clinical standard, first via second-trimester amniocentesis (99.4% to 99.8% accuracy rate) (3) and then, in the 1980s, with first-trimester chorionic villus sampling (CVS) conducted via ultrasound guidance (97.5% to 99.6% accuracy rate) (3). A duration of up to 10 to 14 days for necessary cell cultures was and remains the salient limitation for both amniocentesis and CVS, along with a relatively low-risk of pregnancy loss, which continues to limit this procedure's application among pregnant women younger than 35 years (4). The cfDNA tests rely on DNA fragments shed by the placenta that, in nearly all pregnancies, is equivalent to fetal DNA. These short fragments of DNA, along with fragments from the pregnant woman, float in the woman's blood stream. This mixture of fragments can be extracted and analyzed for extra material from specific chromosomes. In addition to screening for Down syndrome, cfDNA tests can also screen for Edwards syndrome (trisomy 18) and Patau syndrome (trisomy 13), both of which can cause serious developmental and medical issues.
Rapid aneuploidy detection or diagnostic tests, such as fluorescence in situ hybridization (FISH), quantitative fluorescence polymerase chain reaction (QF-PCR), and multiplex ligation-dependent probe amplification (MLPA) has changed the approach to prenatal diagnosis in the last 20 years. FISH uses targeted probes to identify chromosome rearrangements and aneuploidy in interphase nuclei from amniotic fluid and CVS (5). QF-PCR uses fluorescence probes and polymerase chain reaction in a multiplex assay to amplify targeted sections of genome and measure alleles of interest with a fluorescence analyzer. In MLPA the most recent of these techniques, it is not the nucleic acid but the probes added to the samples that are amplified and quantified (5). These techniques offer a number of advantages over traditional cytogenetic testing and have eliminated the need for cell cultures. The resulting time-delay minimization and higher available resolutions continue to influence and alter former referral patterns for genetic testing.
These non-invasive, rapid, and accurate techniques are available to test for the following conditions, which account for more than 80% of known clinically significant prenatal chromosomal abnormalities: trisomy 21, sex chromosome aneuploidies X (Turner syndrome) and XXY (Klinefelter syndrome), autosomal trisomies for chromosome 13 (Patau syndrome) and chromosome 18 (Edwards syndrome), and triploidy. For all these examples, the duration required for detection and diagnosis is 1 to 2 days for FISH and QF-PCR; and 2 to 3 days for MLPA. Most recently, array-based comparative genomic hybridization (CGH) services - known as array CGH or microarrays have become available in clinical laboratories. Microarrays use array CGH to detect gains and losses in genetic material. As a diagnostic tool, microarrays address many of the limitations of current or earlier cytogenetic methods and represent a technological breakthrough (6). In pregnancies with fetal anomalies on ultrasound, array CGH detects 6% additional cytogenomic abnormalities than karyotype alone and 1.7% more even in women of advanced age without fetal anomalies (7). Performing array CGH in cell-free fetal blood is theoretically possible, but at present prenatal array CGH will probably require intact fetal cell(s) derived from CVS or amniocentesis or intact fetal trophoblast from maternal blood (8).
Though prenatal screening tests vary by clinic, the current standard often includes a blood test and an ultrasound during the first trimester. The blood test measures hormone and protein levels that are associated with chromosomal abnormalities. The ultrasound for nuchal translucency test, which measures fluid that accumulates in the back of fetus's neck - it is a characteristic that is strongly associated with an elevated risk of Down syndrome. cfDNA testing is not perfect; however, if blood samples do not include sufficient placental DNA, the results may be inconclusive. And a rare form of Down syndrome called mosaicism, in which abnormalities can vary between the placenta and the fetus can be missed by cfDNA tests. Because of the chance of a false positive, women are advised not to base their decision to terminate their pregnancies on results of cfDNA testing alone. Those who test positive are very strongly urged to undergo an amniocentesis or CVS before taking irreversible action.
Clinical Considerations and Consequences
Chromosomal abnormalities can be devastating, and the overall population risk of a child being born with a congenital abnormality, whether genetically or environmentally determined, is estimated to vary between 3% and 5% (9). Chromosomal abnormalities are observed to occur in 1 in every 156 live births, 1 in 2.5 first-trimester spontaneous abortions, and 1 in 16 neonatal deaths and stillbirths (9). Such odds warrant an abundance of caution in the early application of appropriate diagnostics. Classic clinical indications for prenatal testing include a maternal age of 35 years or older at delivery, abnormal ultrasound findings, abnormal maternal serum screening, a prior child or pregnancy with a chromosomal abnormality, known parental chromosome rearrangement, and family history of a genetic condition. Testing for chromosomal abnormalities can also simply be precautionary, performed to address parental anxiety and/or concerns. We recommend that aneuploidy testing be offered to pregnant women of all ages.
Information to Share with Patients before Prenatal Chromosomal Microarray Analysis:
In addition to the data regarding genetic testing results, the National Institute of Child Health and Human Development (NICHD) study raised several important considerations for the clinical application of chromosomal microarray analysis in the prenatal setting. The potential for detection of clinically uncertain and complicated findings with prenatal chromosomal microarray analysis can result in substantial patient anxiety. This underscores the critical need for comprehensive patient pre-test and post-test genetic counseling from qualified personnel such as a geneticist or genetic counselor about the benefits, limitations, and results of testing so that patients can make informed decisions. Information that should be shared with patients who are considering prenatal chromosomal microarray analysis is provided for use before referral for genetic counseling (30).
- Chromosomal microarray analysis will identify almost all of the abnormalities that are identified by fetal karyotyping and may identify additional specific genetic diseases. It will not identify all genetic disorders.
- Diseases may be identified for which the clinical presentation may vary greatly and range from mild to severe. It may not be possible to predict what the outcome will be in a given patient.
- The test may identify consanguinity (a close blood relationship or incest) or non-paternity.
- Genetic changes may be identified that may or may not cause disease. Samples from both parents may be required to help understand the significance of these results.
- Test results may identify adult-onset diseases that will not affect health during the newborn period or childhood but may have unknown severity later in life. Identification of such findings may also indicate that one of the parents has the same adult-onset disease but has not yet developed symptoms.
Current State of Prenatal Diagnostics
Karyotype analysis, has high diagnostic accuracy and provides a whole-genome view, but has a number of disadvantages. Karyotyping has the limitation of only identifying rearrangements involving >5 to 10 million base pairs, has low-resolution, and has slow turn-around time (up to 10 to 14 days) (5). The advantages associated with FISH analysis include rapid detection, high resolution, and less labor intensity compared with karyotyping. Drawbacks are that analysis is limited to regions targeted by the probes, and therefore, there is need for prior knowledge of the region to be examined.
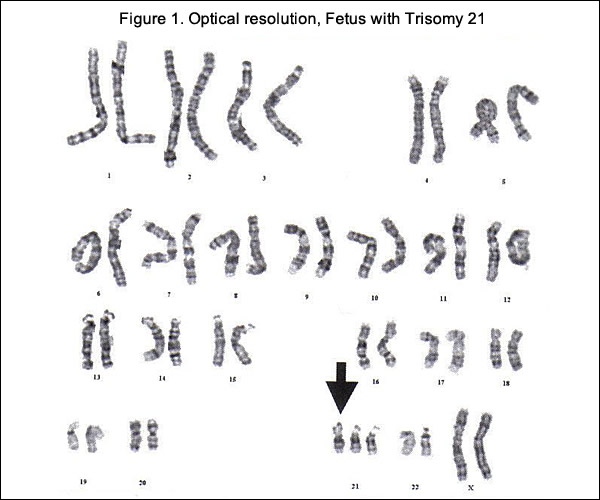
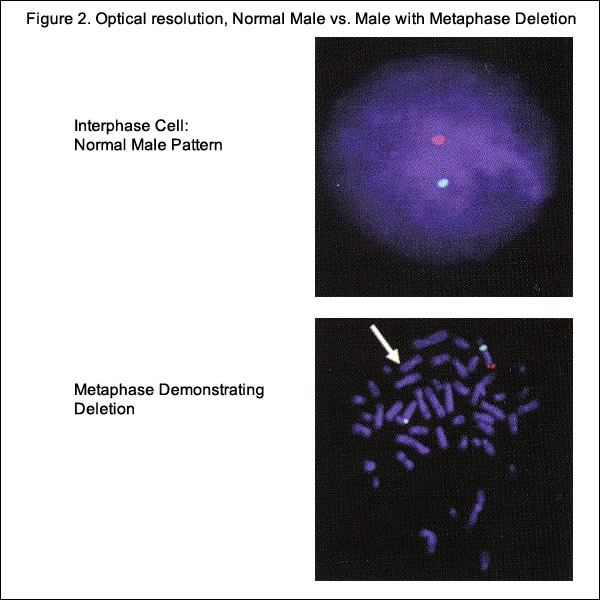
Cell-Free Fetal DNA (cfDNA)
cfDNA fragments are short fragments of DNA found in blood. During pregnancy, there are cfDNA fragments from both the mother and fetus in maternal circulation (14). It is possible to analyze cfDNA to detect common fetal trisomies such as T21, which causes Down syndrome. Fetal trisomy is detected by counting cfDNA fragments. The counting is made possible by using next generation DNA sequencing. Although it has been known for some time that fetal cells enter the maternal circulation, it is still not feasible to sample them adequately in maternal plasma because they are proportionally scarce. However, when fragments of fetal DNA - known as cfDNA were identified in maternal plasma, the equation changed. cfDNA is far more plentiful than fetal cells, and researchers have shown that is specific to the current pregnancy (10). By analyzing cfDNA from maternal plasma, it is now possible to screen for trisomies with high sensitivity and specificity. Early attempts to detect trisomic fetuses using cfDNA required the use of multiple placental DNA or RNA markers, which made the screening test time consuming and expensive (11). Recently, a number of groups have validated a technology known as massively parallel genomic sequencing, which uses a highly sensitive assay to quantify millions of DNA fragments in biological samples in a span of days and has been reported to accurately detect trisomy 13, trisomy 18 and trisomy 21 as early as the 10th week of pregnancy with results available approximately 1 week after maternal sampling (12). It does not replace the precision obtained with diagnostic tests, such as CVS or amniocentesis, and currently does not offer other genetic information.
Indications for Considering the Use of cell-free DNA (cfDNA) (13):
- Maternal age 35 years or older at delivery.
- Fetal ultrasonographic findings indicating an increased risk of aneuploidy.
- History of prior pregnancy with a trisomy.
- Positive test result for aneuploidy, including first trimester, sequential, or integrated screen, or a quadruple screen.
- Parental balanced Robertsonian translocation with increased risk of fetal trisomy or trisomy 21.
Detecting fetal trisomy from the 5% or more of cfDNA in maternal blood that is fetal in origin is more difficult than detecting paternally derived single-gene disorders. Unlike detecting the qualitative differences, trisomy detection requires distinguishing quantitative DNA differences between affected (trisomies) and unaffected (euploid) pregnancies because, of course, both mother and fetus have chromosome 21 DNA sequences. Differentiating trisomic from normal pregnancies is still possible, however, because a pregnancy with a trisomy 21 fetus has more chromosome 21 DNA sequences than one with a normal fetus. The trisomic fetus has three 21 chromosomes, rather than the usual two. The small but finite quantitative differences in the total number of chromosome 21 sequences in maternal blood should be detectable. Given that the fetus is the source of perhaps 5% of cfDNA in maternal plasma, blood from a mother carrying a trisomy 21 pregnancy should have 2.5% more chromosome 21 sequences than if her fetus were not trisomic. This small but finite difference serves as the basis for detecting fetal trisomy (aneuploidy) by analysis of maternal blood. Two general approaches are possible.
Massively Parallel Genomic Sequencing (MPGS)
One technology used in the United States by Sequenom (San Diego, California) and Verinata Health (Redwood City, California) is massively parallel genomic sequencing (MPGS). In MPGS, all cfDNA (maternal plus fetal) is sequenced. From among some 25 million sequences (reads), those relevant to the chromosome being assessed 21, 13, and 18 are quantitatively compared to number of "reads" expected. Several reports validated a threshold using archived or accumulated samples of maternal plasma. In 7% of 576 samples studied in 2011, informative results were not possible, yet all 86 trisomy 21 cases in the remaining samples were successfully detected (15). Another study in 2011 reported detection of 39 of 39 trisomy samples derived from 449 analyzable maternal plasma samples (16). There were no false positives.
In 2012 at Verinata scientists detected 89 of 89 trisomy 21 pregnancies in analyzable samples (17). Of these samples, 3% had insufficient cfDNA. Their method detected 35 of 36 trisomy 18 cases and 11 of 14 trisomy 13 cases. There were no false positive cases for these autosomal trisomies. Also in 2012, researchers in Germany reported on successful use of MPGS of DNA using the technique previously described by Chiu (15) to identify correctly fetal trisomy 21 in all 8 samples from women carrying affected fetuses out of a total of 42 samples. In some publications, investigators reported efforts to optimize quantification of trisomy 21 sequences with new algorithms for calculation of the z score (18). These studies provide confidence that cfDNA can be exploited to detect trisomy 21 pregnancies using a quantitative threshold model. Using MPGS, detection rates are less than 100% for trisomies and 18 but are still very high. In this study MPGS provided an accurate way of detecting the most prevalent aneuploidies associated with cystic hygroma (25). It could advance prenatal care by providing alternative point-of-care non-invasive testing for pregnant women who either decline or do not have access to an invasive procedure.
Targeted DNA Sequencing
Others are using a different approach for detecting fetal trisomies: targeted DNA sequencing. Rather than accumulating and sequencing maternal and fetal cfDNA from all chromosomes, only sequences from those chromosomes necessary for diagnosis are recovered and quantified - chromosome 21, 13 and 18, for example. One can retrieve selected sequences using probes. A set of trisomy 21 cases and controls first yielded 100% detection of trisomic samples (8). In the latest report using this approach, "routinely screened" first-trimester samples were assessed in women of maternal age of 31.8 years (19). The non-invasive rate was again near 5%. Eight of eight trisomy 21 cases and 2 of 3 trisomy 18 cases were detected. The false-positive rate was 0.1%.
A potential advantage of this approach is applicability using smaller percentages of cfDNA. In the initial report, the non-informative rate was 12.6%, but the decision of whether to call a sample informative was based on the ability to obtain information on all 5 chromosomes, not the fewer number in other studies. The non-informative rate for the 3 autosomal trisomies alone (13),(18),(21) is comparable to other reports. Zimmerman and associates studied 116 samples using this approach, targeting 11,000 parental single nucleotide polymorphisms on chromosomes 13, 18, 21, X, and Y (20). The achieved samples studied included 2 trisomy 13 cases; 3 trisomy 18 cases; 11 trisomy 21 cases; two 45,X cases; and two 47,XXY cases. The correct chromosome number was reported in all cases.
Appropriate Clinical Application
Despite the complex background of cfDNA testing, benefits for implementation into patient care clearly exist. First-trimester screening with nuchal translucency and maternal serum screening carries added benefits, including identification of aneuploidies beyond those currently detectable with non-invasive prenatal testing. In a recent publication on non-invasive prenatal testing in an average risk population of women undergoing first-trimester screening, non-invasive prenatal testing detected 55% of the total chromosomal abnormalities (eight of eight cases of trisomy 21 and two of three cases of trisomy 18), whereas first-trimester screening detected 100% (all T21 and T18 as well as seven other deletions, duplications, and other anomalies) (19). In addition, screening for neural tube and ventral wall defects is an important component of current screening protocols. Optimal introduction of non-invasive prenatal testing would preserve the principle of providing prenatal screening and diagnosis in a way that maximizes women's reproductive choice and in addition, ensure that non-invasive prenatal testing is not performed on women who are unaware of the purpose and scope of the test (21).
Counseling regarding the limitations of cfDNA testing should include a discussion that the screening test provides information regarding only trisomy 21 and trisomy 18, in some laboratories trisomy 13. It does not replace the precision obtained with diagnostic tests, such as CVS or amniocentesis, and currently does not offer other genetic information. Other limitations of cfDNA include the lack of outcome data for low-risk populations; therefore, cfDNA testing is not currently recommended for low-risk women. Preliminary data available on twins demonstrate accuracy in a very small cohort, but more information is needed before use of this test can be recommended in multiple gestations (22). In a small percentage of cases, a cfDNA result will not be able to be obtained. To offer cfDNA test, pretest counseling regarding these limitations is recommended. The use of a cfDNA test should be an active, informed choice and not part of routine prenatal laboratory testing. The family history should be obtained to determine if the patient should be offered other forms of screening or prenatal diagnosis for a particular disorder. A baseline ultrasound examination may be useful to confirm viability, a singleton gestation, gestational dating, as well as to rule out obvious anomalies. Referral for genetic counseling is suggested for pregnant patients with positive test results. Because false-positive test results can occur, confirmation with amniocentesis or CVS is recommended. Patients also need to be aware that a negative test result does not ensure an unaffected pregnancy; false-negative test results can occur as well. In this high-risk population, a second-trimester ultrasound examination is suggested to evaluate pregnancies for structural anomalies. In patients in whom a structural fetal anomaly is identified, invasive diagnostic test should be offered because a cfDNA can only detect trisomy 13, trisomy 18, and trisomy 21. Maternal serum alpha-fetoprotein screening or ultrasonographic evaluation for open fetal defects should continue to be offered.
Recommendations and Considerations
Developing reasonable clinical management guidelines and education is essential as the testing becomes more sophisticated. Results of non-invasive prenatal testing can take 1 to 3 weeks, the implications of the turnaround time, especially if a test result is ultimately not provided, need to be considered, especially for patients who are later in the second trimester, for whom it may be too late to pursue other screening or diagnostic testing options. Our suggestions include:
- Patients at increased risk of aneuploidy can be offered testing with cfDNA. This technology can be expected to identify approximately 98% of cases of Down syndrome with a false-positive rate of less than 0.5% (22).
- cfDNA testing should not be part routine prenatal laboratory assessment, but should be an informed patient choice after pretest counseling.
- cfDNA testing should not be offered to low-risk women or women with multiple gestations because it has not been sufficiently evaluated in these groups.
- Pretest counseling should include a review that although the cfDNA is not a diagnostic test, it has high sensitivity and specificity. The test will only screen for the common trisomies, and at the present time, gives no other genetic information about the pregnancy.
- A family history should be obtained before the use of this test to determine if the patient should be offered other forms of screening or prenatal diagnosis for familial genetic disease.
- If a fetal structural anomaly is identified on ultrasound examination, invasive prenatal diagnosis should be offered.
- A negative cfDNA test result does not ensure an unaffected pregnancy.
- A patient with a positive test result should be referred for genetic counseling and offered invasive prenatal diagnosis for confirmation of test results.
- cfDNA does not replace the accuracy and diagnostic precision of prenatal diagnosis with CVS or amniocentesis, which remain an option for women.
- Additional evaluation of cfDNA testing in clinical practice and a mechanism for the systematic reporting of false-positive and false-negative cases will be important before this test is offered widely to the general population of low-risk obstetric patients. In the meantime, incorporating information about the positive predictive value in pretest counseling and in clinical laboratory reports is recommended (27). These experiences reinforce the importance of offering invasive testing to confirm cfDNA results before prenatal decision-making.
Finally, the implications of failed tests need to be considered. Up to 5% of cases do not generate a result, often as a result of insufficient cfDNA (especially for women with high body mass index). It is unknown at what body mass index cutoff the test is no longer worth attempting and in what circumstances a repeat attempt is worthwhile (23).
Non-informative Results
At least 5% of samples can yield non-informative results. One explanation is insufficient cfDNA, based on a defined percentage of cfDNA. An obvious solution might be repeating the assay 1 week later, but this will not necessarily yield better results because cfDNA levels do not increase with advancing gestational age between 10 and 21 weeks. The other general explanation is assay failure. Quality-control problems may decrease with experience but also could be counter-balanced by problems arising as labs process more samples. Currently, diagnosis can be offered only from 10 weeks' gestation, and there may be insufficient DNA before this. With respect to early detection, it is increasing that correct diagnosis of cystic fibrosis and spinal muscular atrophy was recently reported in 63 consecutive cases by analysis of fetal trophoblasts recovered from maternal blood and "blindly" analyzed in the first trimester concurrent to carrying out CVS results that was acted on clinically. This process involves molecular analysis of individual trophoblasts, which can be isolated on filter beginning at 5 weeks gestation (24).
Cost
Cost is a challenge to widespread use of cfDNA analysis for prenatal diagnosis. One vendor posts a list price approximating $2,700 using MPGS technology but takes into account what an individual insurance carrier allocates. Costs should ideally be comparable to those incurred with maternal serum analysis; however, costs need not be precisely identical given that if the false-positive rate is truly 1% or less, savings would occur as a result of the need to perform fewer invasive procedures.
Future of Fetal Chromosomal Testing
Initial reports in 2012 and 2013 are allowing us to conclude confidently that cfDNA analysis is valid for detecting trisomy 21. Detection to date is near 100%. The false-positive is less than 1%, far lower than the 5% expected with maternal serum analysis / neural-tube screening. Fewer invasive procedures will thus be needed. If results continue to be so favorable, there will be much to recommend cfDNA as the primary screening approach for fetal aneuploidy. Initially, guidelines unavoidably will recommend this approach for "high-risk" populations, given reports primarily having involved such samples. A strong case can be made to extend this test to the entire obstetrical population.
Array comparative genome hybridization (CGH) can increase greatly the number of detectable disorders beyond fetal trisomy. Is it then still appropriate to detect only trisomies when array CGH can detect much more? At what cost should an invasive procedure be eschewed? Will the detection of greater numbers of disorders by array CGH (or sequencing the entire fetal genome) trump the non-invasive approach of cfDNA, thus actually generating more invasive test? Or can cfDNA evolve to achieve most of the diagnoses now possible only by array CGH? On the other hand, if intact fetal trophoblasts can be recovered readily, would this be a more attractive option than studying cfDNA unavoidably admixed with maternal DNA? The prenatal landscape is indeed likely to continue to change.
Metabolomic Analysis for First-Trimester Down Syndrome Prediction
Metabolites are low-molecular-weight organic and inorganic chemicals that are the substrates, intermediates, and by-products of enzyme-mediated biochemical reactions in the cell. Metabolomics refers to the systematic identification and quantification of these small (less than 1,500 Da) molecules. Metabolomics may have the potential to provide more fundamental and global information than genomics, proteomics, and transcriptomics and to more precisely reflect the phenotype of the cell. One particular application has been in the development of biomarkers for disparate and complex disorders such as schizophrenia, meningitis, and colon cancer. This a case-control study of pregnancies between 11+0 and 13+6 weeks was drawn from a large prospective study to identify potential biomarkers of pregnancy complications in women attending their first hospital visit at King's College Hospital (London, UK) from March 2003 through February 2009 (26). There were 30 Down syndrome cases and 60 controls in which first-trimester maternal serum was analyzed. Nuclear magnetic resonance-based metabolomic analysis was performed for Down syndrome prediction. Concentrations of 11 metabolites were significantly different in the serum of Down syndrome pregnancies. The combination of 3-hydroxyisovalerate, 3-hydroxybuterate, and maternal age has a 51.9% sensitivity at 1.9% false-positive rate for Down syndrome detection. One multi-marker algorithm had 70% sensitivity at 1.7% false-positive rate. Novel markers such as 3-hydroxybutyrate, involved in brain growth and myelination, and 2-hydroxybutyrate, involved in the defense against oxidative stress, were found to be abnormal (26).
The finding of a profound alternation of the maternal serum metabolome in first-trimester Down syndrome pregnancies is not surprising. The disturbance in trophoblast function leading to changes in the maternal serum concentrations of glycoproteins such as human chorionic gonadotropin and pregnancy-associated plasma protein A is well known. The association between 2-hydroxybutyrate (an organic acid that is elevated in states of oxidative stress), oxidative stress, and single carbon metabolism is interesting in the context of brain dysfunction in Down syndrome. Oxidative stress is considered one of the most likely causes of neurotoxicity in Down syndrome. This preliminary analysis of the metabolomic profile of maternal serum in Down syndrome pregnancies suggests a metabolic environment conducive to a disturbance in fetal central nervous system development. This could lead to the development of additional biochemical markers for the disorder. Some of the metabolites are associated with oxidative stress, poor myelination, and neurotoxicity of the brain in Down syndrome individuals. Metabolomic analysis could lead the development of additional biochemical markers to improve Down syndrome screening. Metabolomics could improve our understanding of the pathologic consequences of the Down syndrome genotype on fetal brain development and potentially lead to prenatal therapy.
Four Fetal Cell-Free DNA Tests Available in the United States
- Harmony Prenatal Test: Ariosa Diagnostics' Harmony Prenatal Test screens for Down syndrome, Patau syndrome, and Edwards syndrome. The test includes an optional analysis of sex chromosome conditions such as Klinefelter syndrome and Turner syndrome. Sex chromosome conditions occur when there is a missing, extra, or incomplete copy of one of the sex chromosomes. It detects >99% of fetal trisomy 21 cases at a false positive rate of <0.1% (28). Optional X and Y chromosome analysis available for fetal sex and X, Y sex chromosome analysis. This test does not assess risk for mosaicism, partial trisomies or translocations. This test is available for all singleton and twin pregnancies, including those conceived by in-vitro fertilization (IVF). A minimum amount of fetal cfDNA is necessary for reliable testing and quality results. It incorporates the measurement of fetal cfDNA into the analysis of every sample. It uses directed analysis for accurate trisomy detection.
Advantages of Directed Analysis:
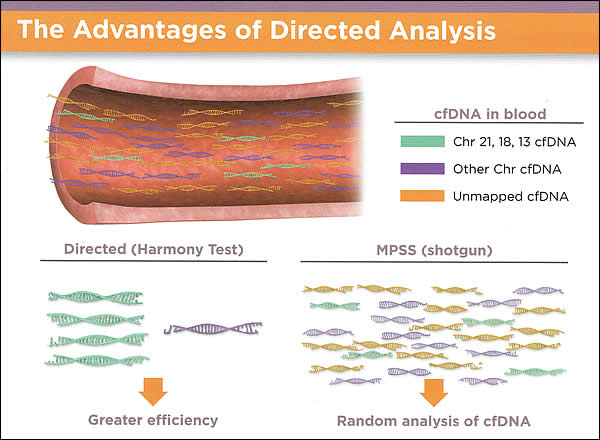
Harmony Prenatal Test uses direct analysis for trisomy detection; blood test to assess the risk of fetal
trisomies and evaluate the X and Y chromosome - MaterniT21 PLUS: Sequenom's MaternalT21 PLUS test screens for Down syndrome, Patau syndrome, Edwards syndrome, sex chromosome conditions, and other more rare conditions. It is recommended that carrier screening for cystic fibrosis (CF), Canavan disease, familial dysautonomia, and Tay-Sachs disease be offered to all women who are pregnant or considering pregnancy who are of Ashkenazi Jewish descent. CF screening is recognized as a benchmark for genetic carrier testing. A buccal swab or blood draw is all that is required. CF genetic screening is the cornerstone of this test and it is most common lethal genetic disease, impacting families of all ethnicities. With nearly 2,000 known mutations in the gene responsible for CF (CFTR), only mutations found to be phenotypically relevant are reported. Spinal muscular atrophy is a severe neuromuscular disease and a leading cause of infant mortality. It is caused by mutations in the SMN1 gene and characterized by degeneration of motor neurons in the spinal cord. Fragile X syndrome is the most common inherited form of intellectual disability, affecting approximately one in 4,000 males and one in 8,000 females from a variety of ethnic backgrounds. Intellectual disability ranges from borderline to severe, although most patients have moderate degrees of mental retardation. Other variable clinical symptoms can be seen with Fragile X. Fragile X syndrome is caused by mutations to the FMR1 gene on the X chromosome and follows an X-linked inheritance pattern.
- Panorama Prenatal Test: Natera's Panorama test screens for Down syndrome, Patau syndrome, and Edwards syndrome. It also screens for sex chromosome conditions and triploidy, a rare chromosomal abnormality in which a fetus has an extra set of chromosomes.
- Verifi Prenatal Test: the basic version of Illumina's Verifi test screens for Down syndrome, Patau syndrome, and Edwards syndrome. A wider option is also available to screen for sex chromosome conditions.
The identification of cfDNA in maternal plasma has led to the introduction of non-invasive prenatal tests with high sensitivity and high specificity for common aneuploidies (trisomy 13, trisomy 18, trisomy 21). A new expanded non-invasive prenatal testing panel that includes five microdeletion syndromes (22q11 deletion syndrome, cri-du-chat [5p minus], Prader Willi or Angelman syndrome, 1p36 deletion syndrome) and two aneuploidies usually associated with nonviable pregnancies (trisomy 16 and trisomy 22) is now available (29). This expanded panel will be performed unless and opt-out box is checked. Because these disorders are so rare, the positive predictive value is expected to be low. As with all new screening tests and technologies, the expanded panel should be appropriately studied before it is widely adopted. At this time, standard of care for diagnosis of microdeletion syndromes remains diagnostic testing (chorionic villus sampling or amniocentesis) followed by fluorescent in situ hybridization or chromosomal microarray. Clinical management guidelines and education are essential. As with all new screening tests and technologies, the expanded panel should be appropriately studied before it replaces current standard of care and changes clinical practice.
Summary
The science of prenatal genetic diagnosis is moving at a place that would have seemed unimaginable just a few years ago. Non-invasive prenatal testing that uses cell-free fetal DNA (cfDNA) from the plasma of pregnant women offers tremendous potential as a screening tool for fetal aneuploidy. Recently, this analysis has become clinically available for women at increased risk of fetal aneuploidy. Recently, a number of groups have validated a technology known as massively parallel genomic sequencing, which uses a highly sensitive assay to quantify millions of DNA fragments in biological samples in a span of days and has been reported to accurately detect trisomy 13, trisomy 18 and trisomy 21 as early as the 10th week of pregnancy with results available approximately 1 week after maternal sampling. cfDNA has a very high detection rate for trisomy 21: 99% or 100%. It does not replace the precision obtained with diagnostic tests, such as CVS or amniocentesis, and currently does not offer other genetic information. Given that the fetus is the source of perhaps 5% of cfDNA in maternal plasma, blood from a mother carrying a trisomy 21 pregnancy should have 2.5% more chromosome 21 sequences than if her fetus were not trisomic.
The introduction of non-invasive prenatal testing as a new high-performance test for universal screening will complement, rather than replace, the ultrasound examination performed at 11 to 13 weeks. The main limiting factor for widespread application of this test in universal screening for trisomies 21 and 18 in all singleton pregnancies at present is cost. To offer a cfDNA test, pretest counseling regarding the limitations is recommended. The use of cfDNA test should be an active, informed choice and not part of routine prenatal laboratory testing. The family history should be reviewed to determine if the patient should be offered other forms of screening or prenatal diagnosis for a particular disorder. cfDNA analysis will remain a screen, not a test requiring no additional assays before a management decision. Expert patient counseling may be important before and after testing. New reports suggest novel metabolomic markers for the first-trimester can predict the presence of fetal Down syndrome. Metabolomics provide insights into the cellular dysfunction in Down syndrome.
References
- Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350(9076):485-487
- Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of human fetus. Sci Transl Med 2012;4(137):137ra76
- ACOG Committee Opinion No. 446: Array comparative genomic hybridization in prenatal diagnosis. Obstet Gynecol 2009;114(5):1161-1163
- Fridman JM. High resolution array genomic hybridization in prenatal diagnosis. Prenat Diagn 2009;29(1):20-28
- Shaffer LG, Bui TH. Molecular cytogenetic and rapid aneuploidy detection methods in prenatal diagnosis. Am J Med Genet C Semin Med Genet 2007;145C(1):87-98
- Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012;367(23):2175-2184
- Mouawia H, Saker A, Jais JP, et al. Circulating trophoblastic cells provide genetic diagnosis in 63 fetuses at risk for cystic fibrosis or spinal muscular dystrophy. Reprod Biomed Online 2012;25(5):508-520
- Ashoor G, Syngelaki A, Wagner M, et al. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol 2012;206(4):319.e1-319.e9
- South ST, Chen Z, Bronthman AR. Genomic medicine in prenatal diagnosis. Clin Obstet Gynecol 2008;51(1):62-73
- Lo YM, Zhang J, Leung TN, et al. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 1999;64(1):218-224
- Papageorgiou EA, Karagrigoriou A, Tsaliki E. et al. Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21. Nat Med 2011;17:510-513
- Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol 2011;204:205.e1-205.e.11
- ACOG Committee Opinion No. 545: Noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol 2012;120(6):1532-1534
- Chiu RWK, Lo D. Noninvasive prenatal diagnosis by fetal nucleic acid analysis in maternal plasma: the coming of age. Semin Fetal Neonatal Med 2011;16:88-93
- Chiu RW, Akolekar R, Zheng YW, et al. Noninvasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 2011;342:c7401
- Ehrich M, Deciu C, Zwieffelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing DNA in maternal blood: a study in clinical setting. Am J Obstet Gynecol 2011;204(3):205.e1-205.e11
- Bianchi DW, Platt LD, Goldberg JD, et al. Maternal Blood is Source to Accurately Diagnose Fetal Aneuploidy (MELISSA) Study Group. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol 2012;119(5):890-901
- Stumm M, Entezami M, Trunk N, et al. Noninvasive prenatal detection of chromosomal aneuploidies karyotyping using different next generation sequencing strategies and algorithms. Prenat Diagn 2012;32(6):569-577
- Nicoladies KH, Syngelaki A, Ashoor G, et al. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am J Obstet Gynecol 2012;207(5):374.e1-374.e6
- Zimmermann B, Hill M, Gemelos G, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat Diagn 2012;32:1-9
- Benn PA, Chapman AR, Ethical challenges in providing noninvasive prenatal diagnosis. Curr Opin Obstet Gynecol 2010;22:128-134
- ACOG Committee Opinion No. 545; December 2012. Noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol 2012;120:1532-1534
- Norton Me, Rose NC, Benn P. Noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol 2013;121:847-850
- Brar H, Wang, E, Struble C, et al. The fetal fraction of cell-free DNA in maternal plasma is not affected by a prior risk of fetal trisomy. J Matern Fetal Neonatal Med 2013;26(2):143-145
- Bianchi DW, Prosen T, Platt LD, et al. Massively parallel sequencing of maternal plasma DNA in 113 cases of fetal nuchal cystic hygroma. Obstet Gynecol 2013;121:1057-1062
- Bahado-Singh RO, Akolekar R, Mandal R, et al. Metabolomic analysis for first-trimester Down syndrome prediction. Am J Obstet Gynecol 2013;208:371.e1-8
- Mennuti MT, Cherry AM, Morrissette JJD, et al. Is it time to sound an alarm about false-positive cell-free DNA testing for fetal aneuploidy? Am J Obstet Gynecol2013;209:415-419
- Norton M, Brar H, Weiss J, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: Results of a Multicenter, Prospective, Cohort Study for Detection of Fetal Trisomy 21 and Trisomy 18. Am J Obstet Gynecol 2012, doi: 10.1016/j. ajog.2012.05.021
- Vora NL, O'Brien BM. Noninvasive prenatal testing for microdeletion syndromes and expanded trisomies. Obstet Gynecol 2014;123:1097-1099
- ACOG Committee Opinion. The use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol 2013;122:1374-1377
نشر: 20 May 2014
Dedicated to Women's and Children's Well-being and Health Care Worldwide
www.womenshealthsection.com


